Computational Scattered Light Imaging
Computational Scattered Light Imaging (ComSLI) allows detailed reconstructions of fiber pathways, also in tissues with densely interwoven fibers which cannot be easily disentangled using standard microscopy or polarization-based techniques. ComSLI has been widely used to resolve complex nerve fiber crossings in brain tissue sections (see image below), but can also be applied to other fibrous tissue structures such as muscle or collagen fibers: The fiber orientations (middle) are obtained by analyzing scattering patterns (right) which reflect the distribution of scattered light behind the sample for each measured point in the tissue section (left).
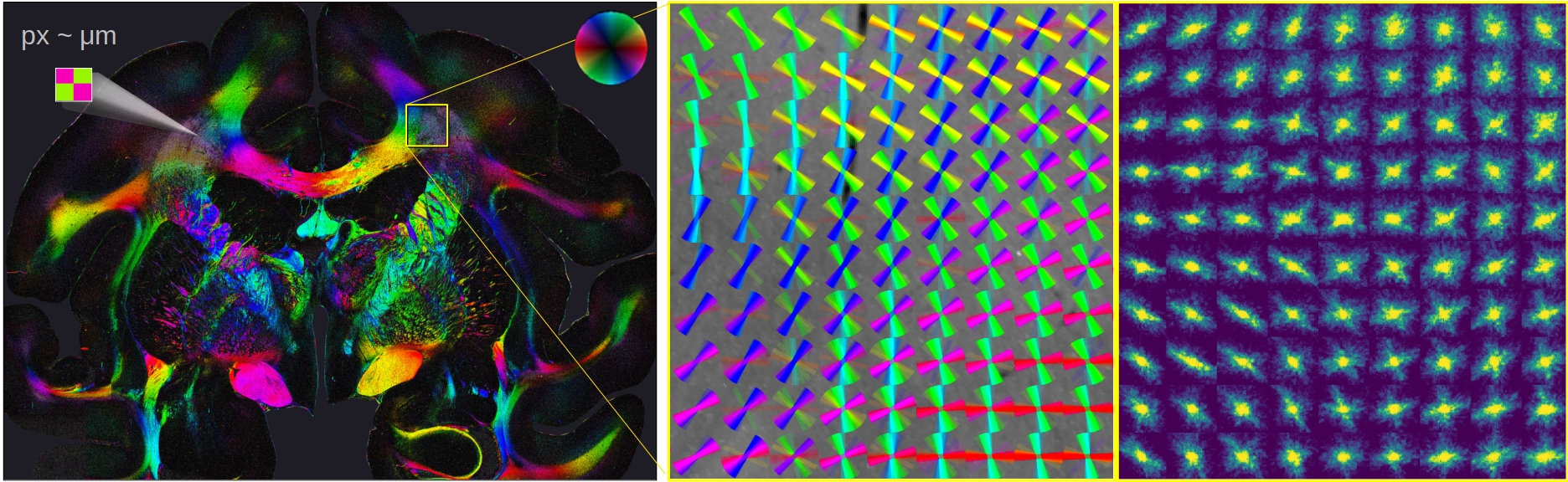
While other scattering techniques require time-consuming raster-scanning of the tissue section and dedicated equipment, ComSLI yields scattering patterns for each image pixel in parallel and can be realized with standard LED light source and camera. As the technique is label-free and uses biologically save levels of illumination, ComSLI can be applied to various tissue types, opening up many new fields of research. In contrast to polarization-based techniques, ComSLI does not rely on birefringence, making it unique for visualizing nerve fiber pathways in paraffin-embedded tissues, which are commonly used in histopathology laboratories and provide little or no birefringence contrast for nerve fibers.
Read More ...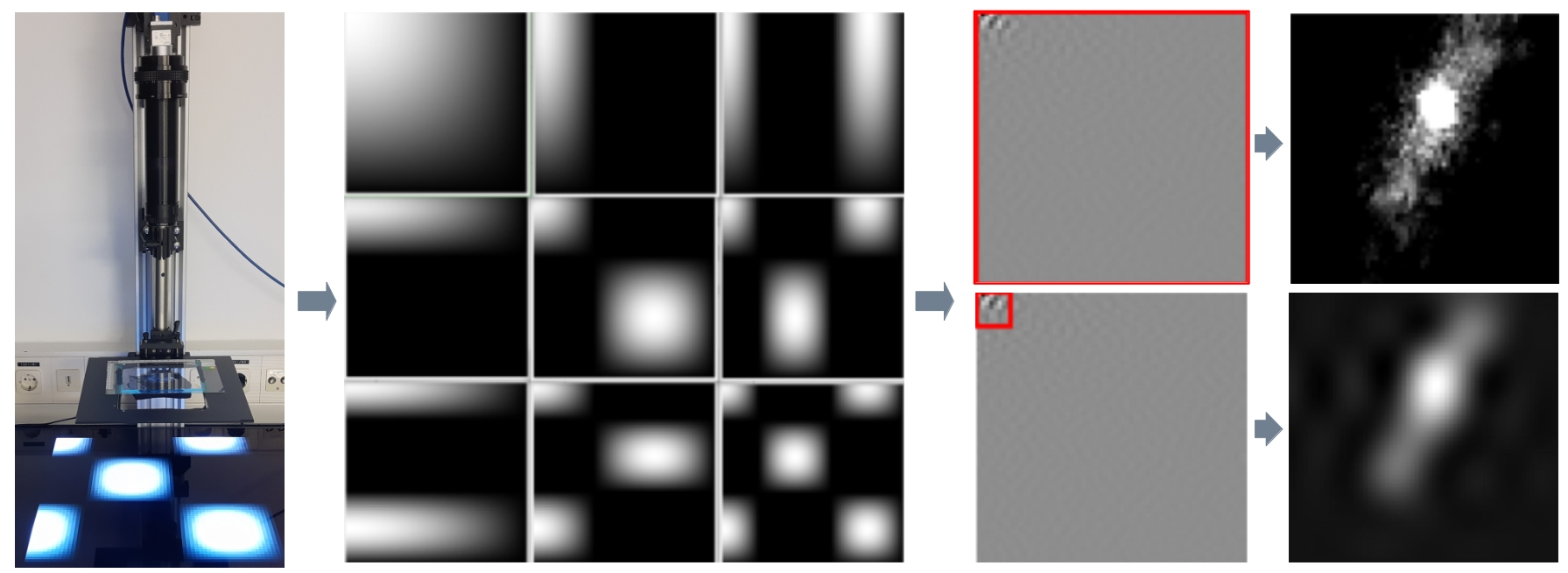
Hardware Development
The current measurement procedure of Computational Scattered Light Imaging can be very time and data consuming, preventing measurements of large volumes. We investigate compressed sensing to realize high-throughput ComSLI measurements, and optimize the light source and illumination patterns.
In addition, we want to further develop the hardware to enable measurements of the tissue block before cutting as well as in-depth scans. Another aim is the miniaturization of the setup to enable clinical applications.
Analysis of Scattering Signals
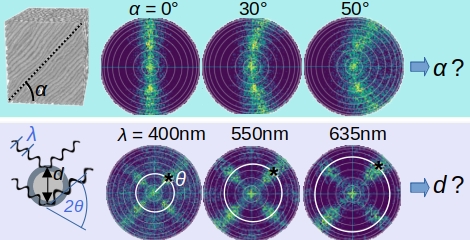
We use advanced signal analysis and machine learning approaches to enhance the interpretation of measured scattering signals, using simulated scattering patterns as training data.
We enhance the analysis of scattering patterns to extract not only in-plane but also out-of-plane fiber orientations. Moreover, we want to explore if scattering patterns obtained from ComSLI measurements with different wavelengths can be used to obtain information about fiber diameters, which are currently only accessible with sophisticated methods like electron or super-resolution microscopy.
Multi-Modal Imaging
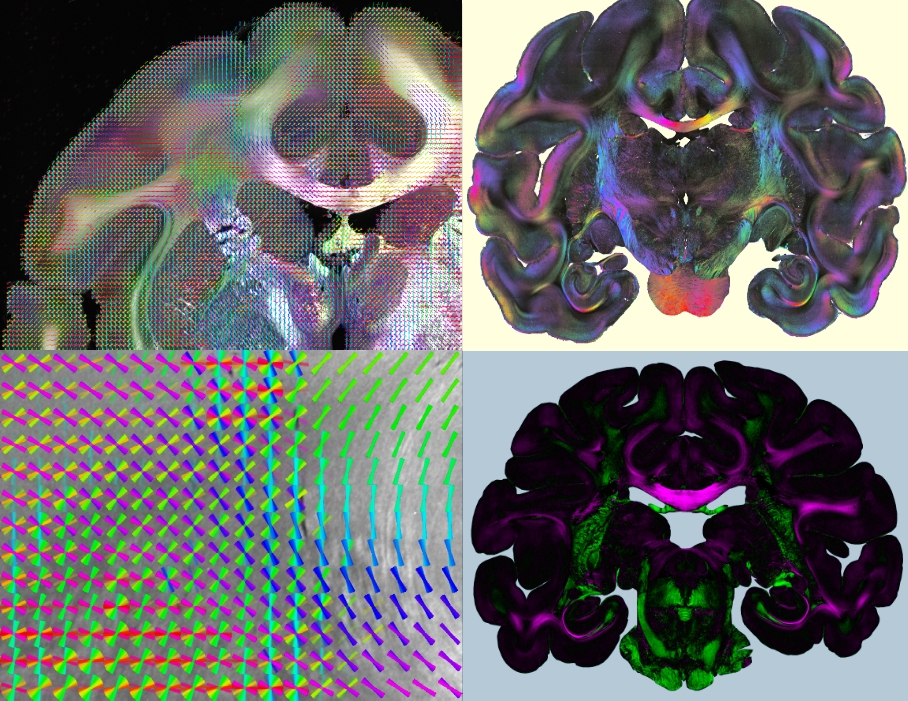
We are building a Scattering Polarimeter to combine polarized and scattered light imaging in one imaging system. In this way, we want to realize fast and fully automated multi-modal imaging of tissue samples without need for image registration or manual work. Apart from hardware development, we work on the generation of multi-modal analysis tools to integrate information from the different imaging techniques.
While Computational Scattered Light Imaging allows to determine multiple nerve fiber orientations for each pixel in the image plane, polarization microscopy provides the three-dimensional orientations of nerve fibers with high accuracy in regions without fiber crossings. As both techniques are compatible and complement each other, a combination of the two is highly beneficial, allowing for multi-modal brain imaging.
Clinical Applications
We explore broader applications of Computational Scattered Light Imaging beyond neuroanatomy, and study different healthy and diseased histological tissues.
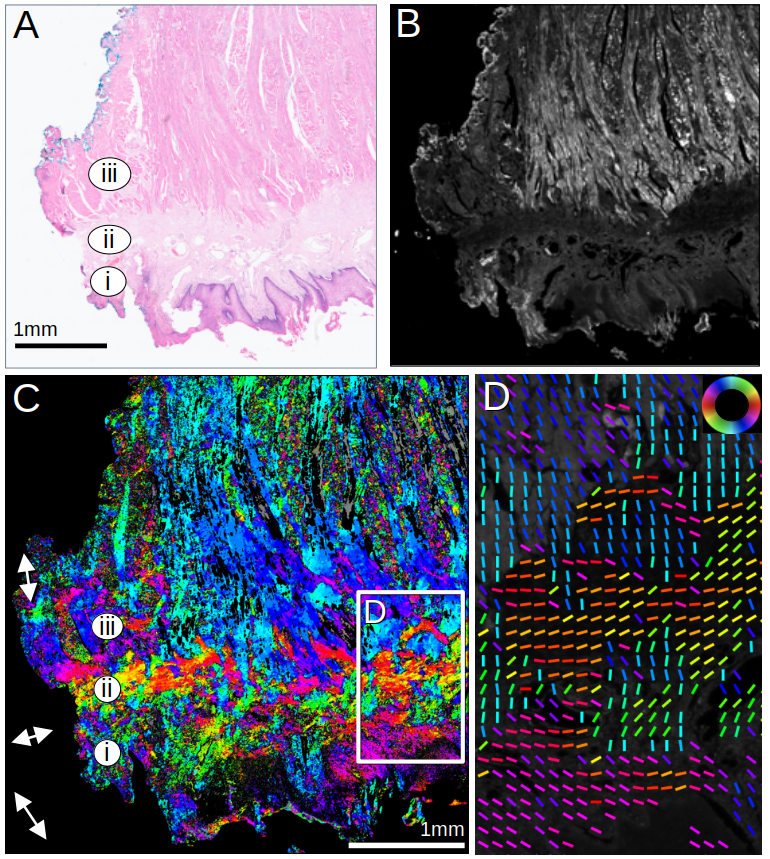
As ComSLI is based on light scattering on micrometer-sized structures, it does not only allow the reconstruction of nerve fiber orientations, but also of other structures with similar dimensions and suitable refractive indices, like muscle or collagen fibers in various tissue types, such as oral and colorectal tissues. The image on the left shows a human tongue muscle measured with ComSLI. (A) Brightfield H&E image; annotated are (i) epithelium, (ii) stroma, and (iii) inner muscle layer. (B) Average scattering signal. (C) ComSLI fiber orientation map, color-coded according to colorwheel in (D). (D) Zoomed-in area from (C) including different fiber layers; fiber orientations are displayed as colored lines for every 25th pixel (for more details, see here).
New collagen fibers begin to grow atop pre-existing fibers in healthy tissue as soon as tumor cells proliferate and eventually metastasize. This growth of new fibrous connective tissue around the tumor nests, known as a desmoplastic reaction, also alters the organization of pre-existing structures within the extracellular matrix (ECM). Studies have demonstrated a correlation between the organization of collagen fibers and patient survival rates. Our research has yielded promising results, showing that ComSLI can effectively visualize changes associated with cancer in muscle and collagen fibers across various cancer types. These findings have significant implications for the potential clinical applications of ComSLI in predicting cancer progression. In collaboration with the Erasmus Medical Center in Rotterdam, we are exploring the use of ComSLI to analyze the fiber architecture of archived histological tissues as a potential prognostic marker.