Computational Scattered Light Imaging
Computational Scattered Light Imaging (ComSLI) allows detailed reconstructions of fiber pathways, also in tissues with densely interwoven fibers which cannot be easily disentangled using standard microscopy or polarization-based techniques. ComSLI has been widely used to resolve complex nerve fiber crossings in brain tissue sections (see image below), but can also be applied to other fibrous tissue structures such as muscle or collagen fibers: The fiber orientations (middle) are obtained by analyzing scattering patterns (right) which reflect the distribution of scattered light behind the sample for each measured point in the tissue section (left).
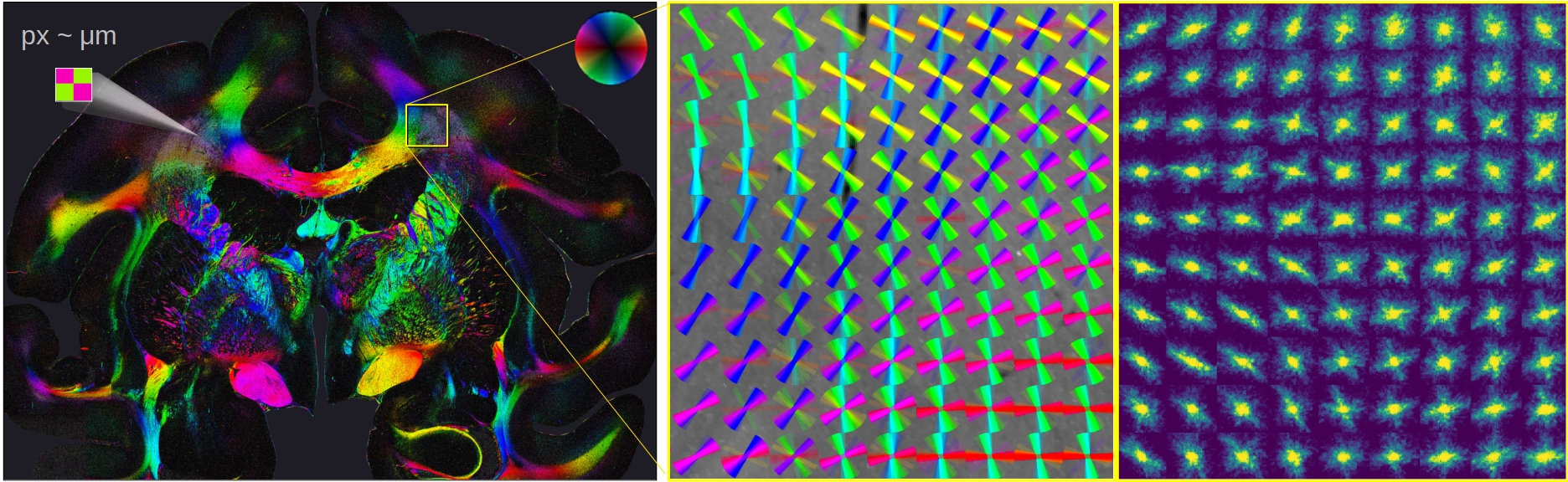
While other scattering techniques require time-consuming raster-scanning of the tissue section and dedicated equipment, ComSLI yields scattering patterns for each image pixel in parallel and can be realized with standard LED light source and camera. As the technique is label-free and uses biologically save levels of illumination, ComSLI can be applied to various tissue types, opening up many new fields of research. In contrast to polarization-based techniques, ComSLI does not rely on birefringence, making it unique for visualizing nerve fiber pathways in paraffin-embedded tissues, which are commonly used in histopathology laboratories and provide little or no birefringence contrast for nerve fibers.
From Simulation to Measurement
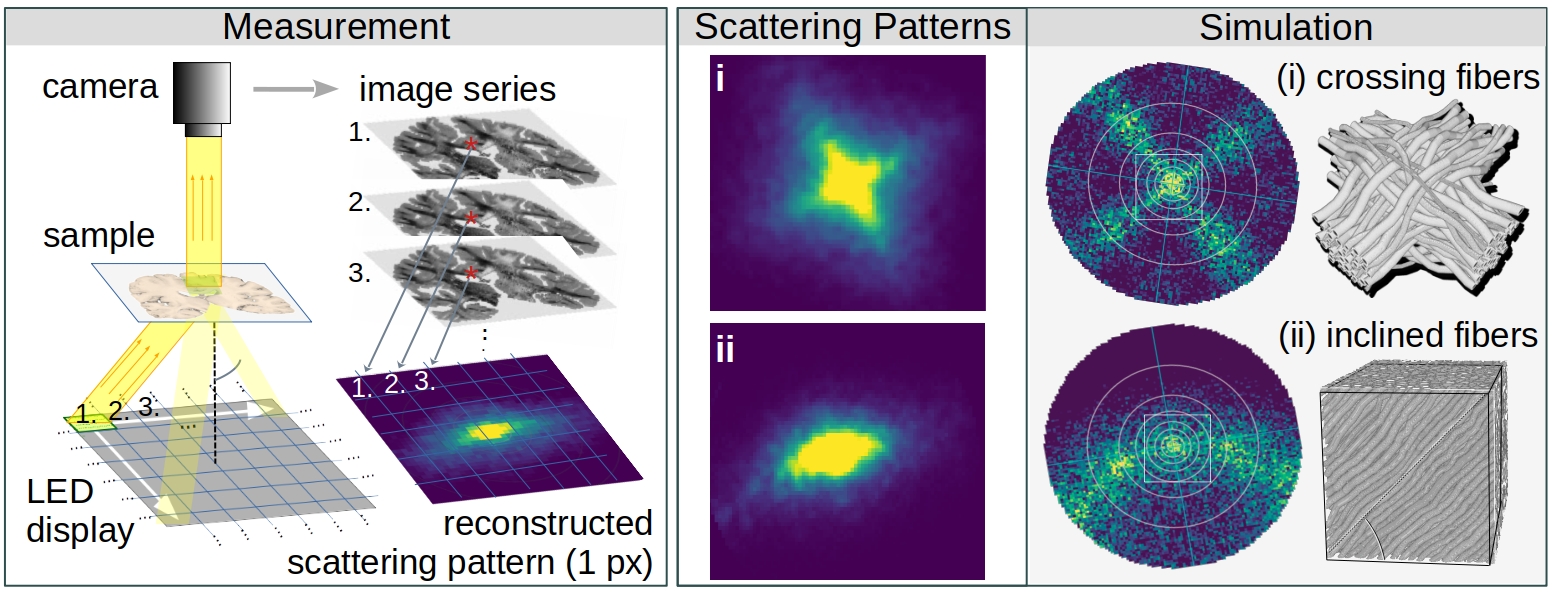
The figure above illustrates the principles of Computational Scattered Light Imaging. We know from simulation studies (right) that the distribution of scattered light behind a sample (scattering pattern) reveals information about its underlying fiber structures, e.g. the individual directions of crossing fibers (i) and the out-of-plane fiber inclination (ii). While other scattering techniques raster-scan the sample with a narrow light beam and measure the scattering pattern behind the sample for each point of illumination, ComSLI uses a reverse setup: The whole field of view is illuminated by light from different angles and an image of the normally through-scattered light is recorded for each angle of illumination (left). In this way, scattering patterns can be obtained for all image pixels (px~μm) in parallel, providing micrometer resolution. The shape and positions of the peaks in the scattering patterns are used to compute the fiber orientations for each image pixel, as illustrated in the image on top. To compute fiber orientations from 1D scattering signals, the SLIX software has been developed.
Further Reading:
- Uncovering microstructural architecture from histology with ComSLI (bioRxiv, 2024)
- Scatterometry measurements with Scattered Light Imaging (Front. Neuroanat., 2021)
- Scattered Light Imaging (NeuroImage, 2021)
- Comparison of light and X-ray scattering and dMRI (eLife, 2023)
- Machine learning approach for improved analysis of scattering patterns (IEEE Proc. 2022)
- Coherent Fourier scatterometry (Biomed. Opt. Express, 2020)
- Light scattering measurements and simulations (Phys. Rev. X, 2020)
- Scattered Light Imaging ToolboX (SLIX): https://github.com/3d-pli/SLIX
In previous puplications, ComSLI is still referred to as Scattered Light Imaging (SLI). We added the term “computational” later to avoid confusion with other scattering techniques and stress the computational aspect.