Bachelor & Master Thesis Projects
We welcome driven students with a background in applied physics, nanobiology, microscopy, or data science who want to work at the interface of computational imaging and biomedical research - either on the experimental side (microscopy, hardware, tissue preparation) or on the computational side (image processing and signal analysis).
If you’re interested in a Master thesis project, please contact Miriam Menzel for possible projects (see below for examples of possible theses).
We currently don’t have any Bachelor thesis projects available.
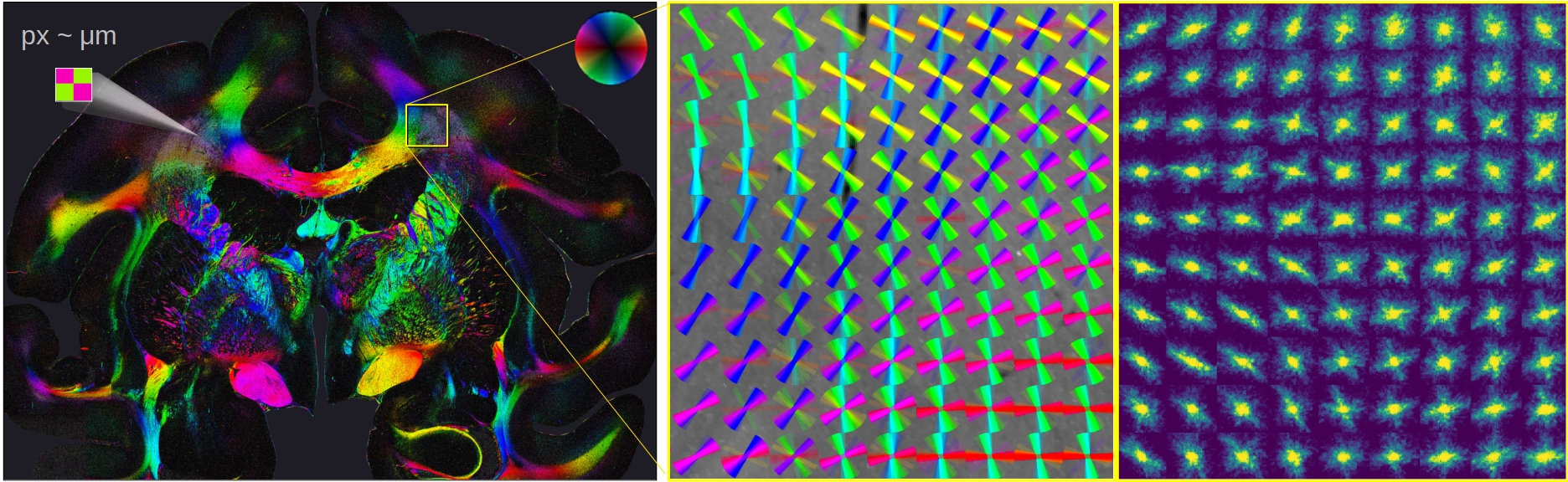
Computational Scattered Light Imaging (ComSLI) resolves nerve fiber pathways and their crossings with micrometer resolution. While other scattering techniques raster-scan the tissue with a light beam and measure the distribution of scattered light behind the sample, ComSLI uses a reverse setup: The whole tissue section is illuminated from many different angles and the normally transmitted light is measured, thus enabling much higher resolutions and requiring only standard optical components (LED light source and camera). This makes ComSLI a highly promising imaging technique, enabling to disentangle complex fiber structures - not only in brain tissue, but also in other biological tissues with comparable fiber structures (e.g. muscle or collagen).
Master Thesis Projects (MEP) - Examples
Please contact Dr. Miriam Menzel if you’re considering to pursue a Master thesis in the Menzel lab, including your list of courses and preferred start/end date. The projects listed below are only examples – we can always find a project tailored to your needs and interests, and we’re flexible with the start date.
Exploiting tissue composition with polarized light scattering
In brain tissue sections, an interesting effect has been observed: Some regions let more light through when the light is polarized parallel to the nerve fibers. Other regions let more light through when the light is polarized perpendicular to the nerve fibers. In this project, we will study to what degree this effect is caused by scattering and how it can be used to distinguish different tissue regions. Apart from brain tissue, we will measure biological tissues with other types of fibers (muscle, collagen) to see if they show similar effects.
Combining fluorescence and scattered light imaging
Computational Scattered Light Imaging allows to distinguish intricately entangled fibers (nerves, collagen, etc.) and their intersections at micrometer scale. Whole-slide fluorescence microscopy enables high-throughput scanning and digitization of an entire microscope slide. It has significant applications in pathology, allowing for rapid screening and analysis of tissue samples. In this project, we will develop an imaging system that combines fluorescence and scattered light imaging.
Analyzing fiber sizes using different wavelengths
It is expected that the scattering of light depends on the feature size relative to the wavelength. In this project, we will systematically compare scattering signals obtained from measurements with different wavelengths on various tissue sections, to better understand how they are related to the underlying fiber structures, and how these measurements can be used to estimate the fiber sizes. Results can be compared to 3D-nanoprinted fiber models with different fiber diameters.
Analyzing different types of fibers
Up to now, Computational Scattered Light Imaging has mostly been used to reconstruct nerve fiber pathways in brain tissue, but it can also be applied to other fibrous structures like muscle or collagen fibers. In this project, we study ComSLI measurements of various biological tissue samples, and compare the scattering signals obtained from nerve, muscle, and collagen fibers. In this way, we want to better understand how these different types of fibers can be distinguished. Another aim is to distinguish tumor from healthy tissue. Depending on your interests, the project can be more software-focused (exploiting machine learning approaches for tissue classification) or experimentally-focused (modifying measurement parameters for enhancing the scattering contrast of non-neuronal structures).
Optimizing illumination patterns for scatterometry measurements
In ComSLI, we usually only measure a single ring of the scattering pattern to determine the in-plane fiber orientations; measuring an entire scattering pattern is time consuming. However, the scattering patterns contain valuable information, e.g., about the out-of-plane orientation of the fibers. In this project, we will explore different illumination patterns and colors to extract the most relevant information of the scattering patterns while keeping the number of illuminations to a minimum.
Confidence map for scattered light imaging measurements
Currently, the measurement results are mostly compared qualitatively, making it difficult to optimize measurements and judge differences between samples. In this project, we will develop a quality measure to better quantify the measured scattering signals and develop a confidence map to indicate the reliability of computed fiber orientations, taking regional information into account.